|
ECG Analysis
Data Set
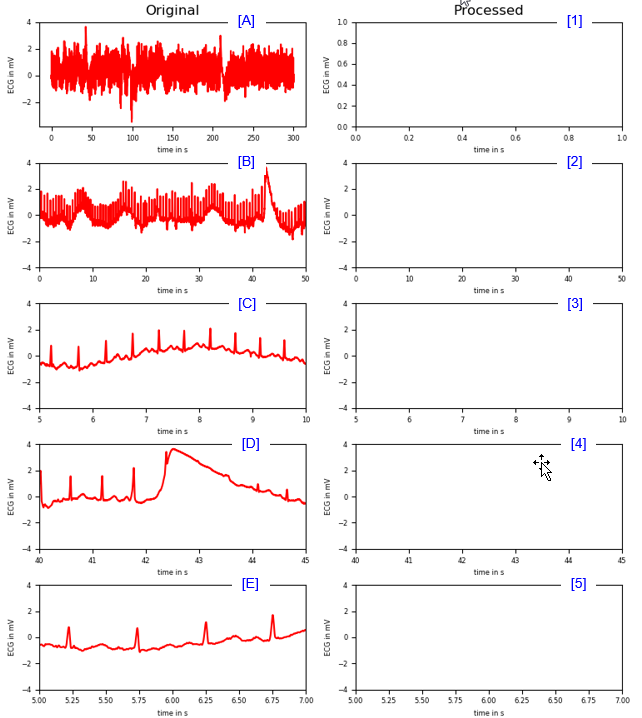
|
import numpy as np
import matplotlib.pyplot as plt
import scipy as sp
import scipy.misc as spm
from scipy.fftpack import fft
from scipy import signal
################################################################
# Data Set #
################################################################
fs = 360;
fn = fs / 2; # Nyquist Frequency
x = spm.electrocardiogram();
t = np.linspace(0,x.size-1,x.size) / fs;
f = np.linspace(0,x.size-1,x.size) * (fs / x.size);
################################################################
# PROCESSING #
################################################################
n = 101;
a = 1;
ftap = signal.firwin(n,cutoff = 0.15, window = "hamming");
xf = signal.lfilter(ftap, a, x);
################################################################
# PLOT #
################################################################
FontSize_Xlabel = 6;
FontSize_Ylabel = 6;
bw = 0.40;
bh = 0.145;
bx = 0.025;
by = 0.1;
bx_gap = 0.075;
by_gap = 0.05;
by_shift = -0.05;
bx_shift = 0.05;
pltRegion1 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 4*(by_gap + bh) + by_shift, bw, bh];
pltRegion2 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 3*(by_gap + bh) + by_shift, bw, bh];
pltRegion3 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 2*(by_gap + bh) + by_shift, bw, bh];
pltRegion4 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 1*(by_gap + bh) + by_shift, bw, bh];
pltRegion5 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 0*(by_gap + bh) + by_shift, bw, bh];
pltRegion6 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 4*(by_gap + bh) + by_shift, bw, bh];
pltRegion7 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 3*(by_gap + bh) + by_shift, bw, bh];
pltRegion8 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 2*(by_gap + bh) + by_shift, bw, bh];
pltRegion9 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 1*(by_gap + bh) + by_shift, bw, bh];
pltRegion10 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 0*(by_gap + bh) + by_shift, bw, bh];
fig = plt.figure();
ax1 = fig.add_axes(pltRegion1);
ax2 = fig.add_axes(pltRegion2);
ax3 = fig.add_axes(pltRegion3);
ax4 = fig.add_axes(pltRegion4);
ax5 = fig.add_axes(pltRegion5);
ax6 = fig.add_axes(pltRegion6);
ax7 = fig.add_axes(pltRegion7);
ax8 = fig.add_axes(pltRegion8);
ax9 = fig.add_axes(pltRegion9);
ax10 = fig.add_axes(pltRegion10);
ax = ax1;
ax.plot(t, x,'r-');
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax.set_title("Original");
ax = ax2;
ax.plot(t, x,'r-');
ax.set_xlim(0.0,50);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax3;
ax.plot(t, x,'r-');
ax.set_xlim(5.0,10.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV');
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax4;
ax.plot(t, x,'r-');
ax.set_xlim(40.0,45.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax5;
ax.plot(t, x,'r-');
ax.set_xlim(5.0,7.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax6;
#ax.plot(t,xf,'b-');
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax.set_title("Processed");
ax = ax7;
#ax.plot(t, xf,'b-');
ax.set_xlim(0.0,50);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax8;
#ax.plot(t, xf,'b-');
ax.set_xlim(5.0,10.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax9;
#ax.plot(t, xf,'b-');
ax.set_xlim(40.0,45.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax10;
#ax.plot(t, xf,'b-');
ax.set_xlim(5.0,7.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
plt.gcf().set_size_inches(8.0,9);
fig.show();
|
Removal of Trends
< High Pass Filter >
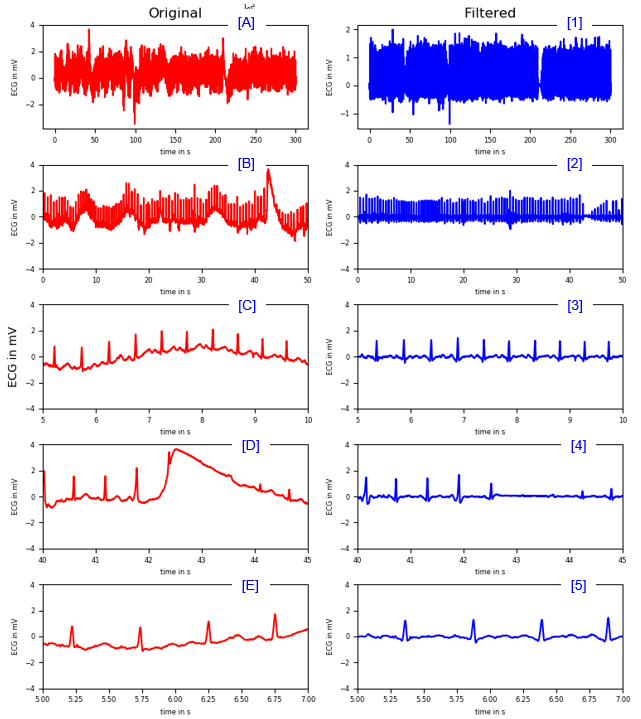
|
################################################################
# PROCESSING #
################################################################
n = 101;
a = 1;
ftap = signal.firwin(n,cutoff = 0.03, window = "hamming",pass_zero = False);
xf = signal.lfilter(ftap, a, x);
|
< Band Stop Filter >
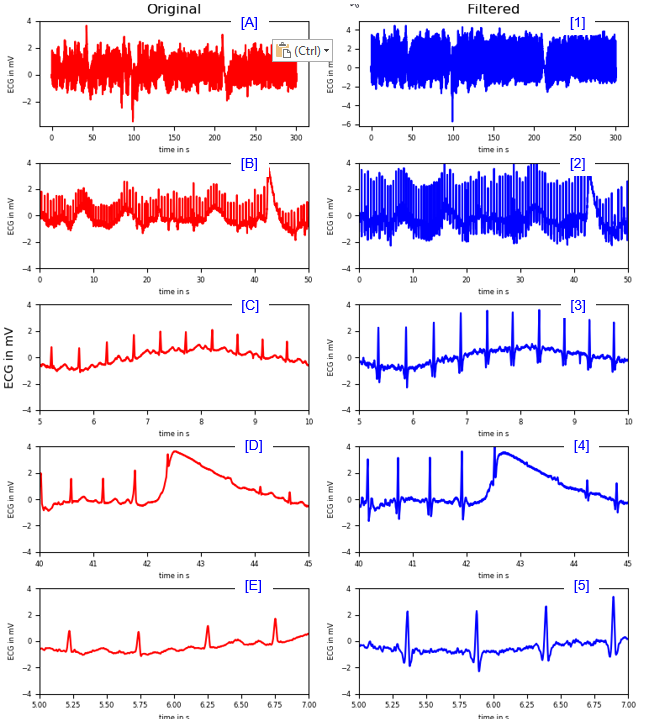
|
################################################################
# PROCESSING #
################################################################
n = 101;
a = 1;
ftap = signal.firwin(n,cutoff = [0.007,0.05], window = "hamming");
xf = signal.lfilter(ftap, a, x);
|
< Band Stop and High Pass Filter >
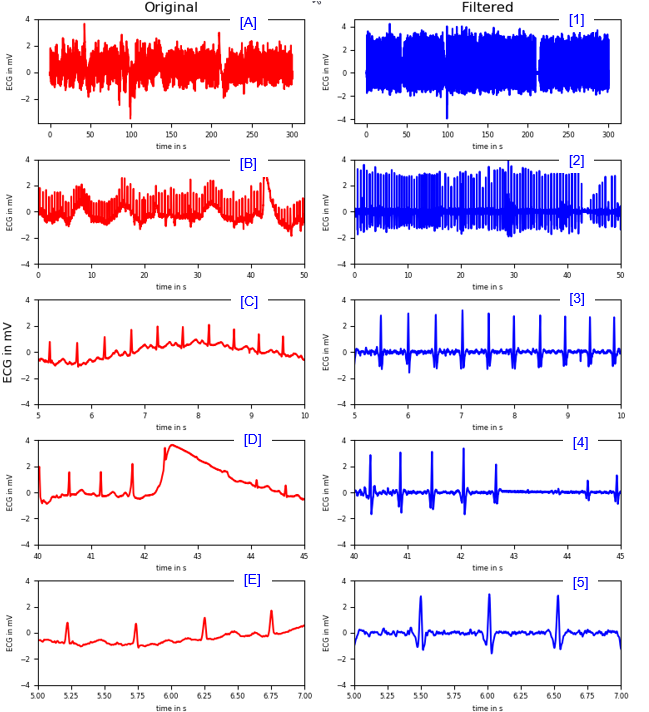
|
################################################################
# PROCESSING #
################################################################
n = 101;
a = 1;
ftap = signal.firwin(n,cutoff = [0.007,0.05], window = "hamming");
xf = signal.lfilter(ftap, a, x);
ftap = signal.firwin(n,cutoff = 0.03, window = "hamming",pass_zero = False);
xf = signal.lfilter(ftap, a, xf);
|
< Band Stop , High Pass and Low Pass Filter >
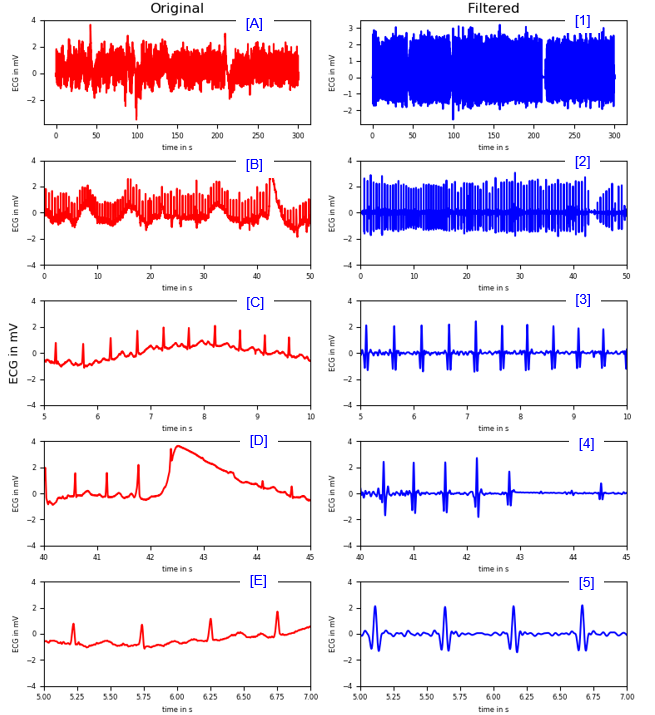
|
################################################################
# PROCESSING #
################################################################
n = 101;
a = 1;
ftap = signal.firwin(n,cutoff = [0.007,0.05], window = "hamming");
xf = signal.lfilter(ftap, a, x);
ftap = signal.firwin(n,cutoff = 0.03, window = "hamming",pass_zero = False);
xf = signal.lfilter(ftap, a, xf);
ftap = signal.firwin(n,cutoff = 0.15, window = "hamming");
xf = signal.lfilter(ftap, a, xf);
|
Finding R peaks
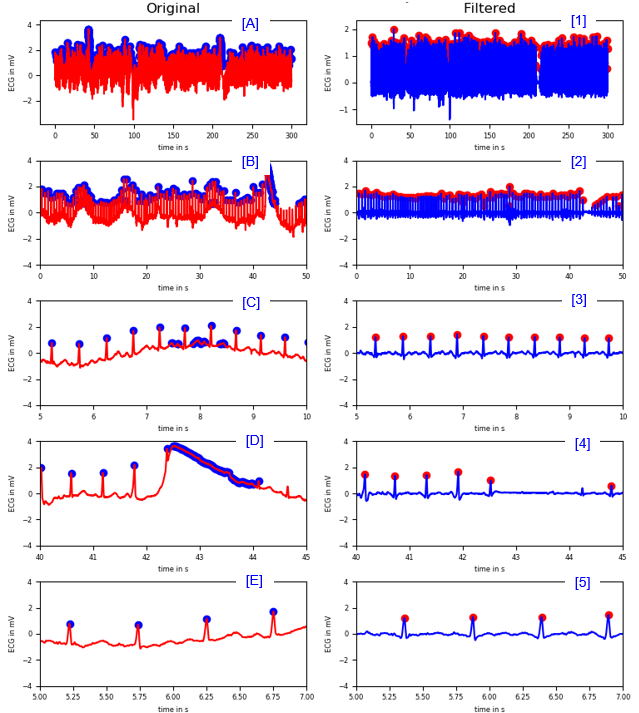
|
import numpy as np
import matplotlib.pyplot as plt
import scipy as sp
import scipy.misc as spm
from scipy.fftpack import fft
from scipy import signal
from scipy.signal import find_peaks
fs = 360;
fn = fs / 2; # Nyquist Frequency
x = spm.electrocardiogram();
t = np.linspace(0,x.size-1,x.size) / fs;
f = np.linspace(0,x.size-1,x.size) * (fs / x.size);
peaks_x,_ = find_peaks(x, height = 0.7);
n = 101;
a = 1;
ftap = signal.firwin(n,cutoff = 0.03, window = "hamming",pass_zero = False);
xf = signal.lfilter(ftap, a, x);
peaks_xf,_ = find_peaks(xf, height = 0.5);
#######################################################
# PLOT #
#######################################################
FontSize_Xlabel = 6;
FontSize_Ylabel = 6;
bw = 0.40;
bh = 0.145;
bx = 0.025;
by = 0.1;
bx_gap = 0.075;
by_gap = 0.05;
by_shift = -0.05;
bx_shift = 0.05;
pltRegion1 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 4*(by_gap + bh) + by_shift, bw, bh];
pltRegion2 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 3*(by_gap + bh) + by_shift, bw, bh];
pltRegion3 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 2*(by_gap + bh) + by_shift, bw, bh];
pltRegion4 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 1*(by_gap + bh) + by_shift, bw, bh];
pltRegion5 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 0*(by_gap + bh) + by_shift, bw, bh];
pltRegion6 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 4*(by_gap + bh) + by_shift, bw, bh];
pltRegion7 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 3*(by_gap + bh) + by_shift, bw, bh];
pltRegion8 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 2*(by_gap + bh) + by_shift, bw, bh];
pltRegion9 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 1*(by_gap + bh) + by_shift, bw, bh];
pltRegion10 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 0*(by_gap + bh) + by_shift, bw, bh];
fig = plt.figure();
ax1 = fig.add_axes(pltRegion1);
ax2 = fig.add_axes(pltRegion2);
ax3 = fig.add_axes(pltRegion3);
ax4 = fig.add_axes(pltRegion4);
ax5 = fig.add_axes(pltRegion5);
ax6 = fig.add_axes(pltRegion6);
ax7 = fig.add_axes(pltRegion7);
ax8 = fig.add_axes(pltRegion8);
ax9 = fig.add_axes(pltRegion9);
ax10 = fig.add_axes(pltRegion10);
ax = ax1;
ax.plot(t, x,'r-');
ax.scatter(t[peaks_x],x[peaks_x],c='blue');
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax.set_title("Original");
ax = ax2;
ax.plot(t, x,'r-');
ax.scatter(t[peaks_x],x[peaks_x],c='blue');
ax.set_xlim(0.0,50);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax3;
ax.plot(t, x,'r-');
ax.scatter(t[peaks_x],x[peaks_x],c='blue');
ax.set_xlim(5.0,10.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax4;
ax.plot(t, x,'r-');
ax.scatter(t[peaks_x],x[peaks_x],c='blue');
ax.set_xlim(40.0,45.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax5;
ax.plot(t, x,'r-');
ax.scatter(t[peaks_x],x[peaks_x],c='blue');
ax.set_xlim(5.0,7.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax6;
ax.plot(t,xf,'b-');
ax.scatter(t[peaks_xf],xf[peaks_xf],c='red');
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax.set_title("Filtered");
ax = ax7;
ax.plot(t, xf,'b-');
ax.scatter(t[peaks_xf],xf[peaks_xf],c='red');
ax.set_xlim(0.0,50);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax8;
ax.plot(t, xf,'b-');
ax.scatter(t[peaks_xf],xf[peaks_xf],c='red');
ax.set_xlim(5.0,10.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax9;
ax.plot(t, xf,'b-');
ax.scatter(t[peaks_xf],xf[peaks_xf],c='red');
ax.set_xlim(40.0,45.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax10;
ax.plot(t, xf,'b-');
ax.scatter(t[peaks_xf],xf[peaks_xf],c='red');
ax.set_xlim(5.0,7.0);
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
plt.gcf().set_size_inches(8.0,9);
fig.show();
|
Finding R peak Interval
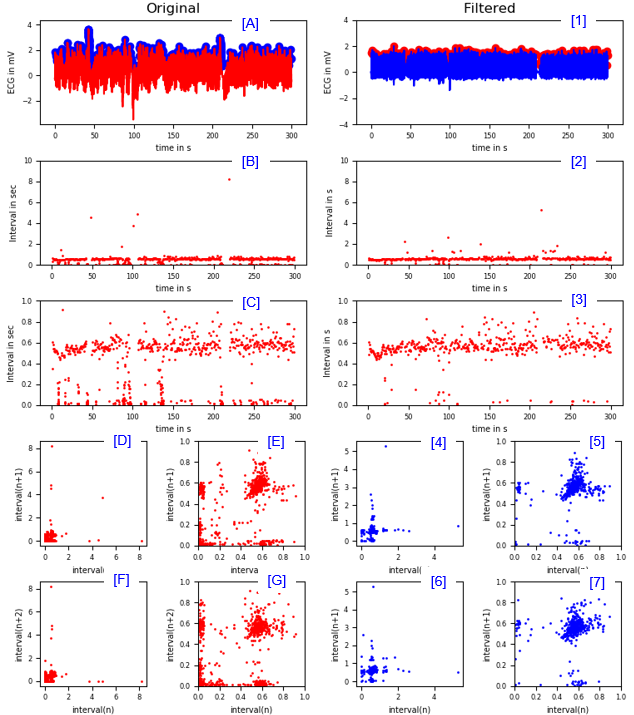
|
import numpy as np
import matplotlib.pyplot as plt
import scipy as sp
import scipy.misc as spm
from scipy.fftpack import fft
from scipy import signal
from scipy.signal import find_peaks
fs = 360;
fn = fs / 2; # Nyquist Frequency
x = spm.electrocardiogram();
t = np.linspace(0,x.size-1,x.size) / fs;
f = np.linspace(0,x.size-1,x.size) * (fs / x.size);
peaks_x,_ = find_peaks(x, height = 0.7);
t_peak_x = t[peaks_x];
w = [1,-1];
x_peak_interval = np.convolve(t_peak_x,w,mode='same');
n = 101;
a = 1;
ftap = signal.firwin(n,cutoff = 0.03, window = "hamming",pass_zero = False);
xf = signal.lfilter(ftap, a, x);
peaks_xf,_ = find_peaks(xf, height = 0.5);
t_peak_xf = t[peaks_xf];
w = [1,-1];
xf_peak_interval = np.convolve(t_peak_xf,w,mode='same');
#######################################################
# PLOT #
#######################################################
FontSize_Xlabel = 7;
FontSize_Ylabel = 7;
bw = 0.40;
bh = 0.145;
bx = 0.025;
by = 0.1;
bx_gap = 0.075;
by_gap = 0.05;
by_shift = -0.05;
bx_shift = 0.05;
pltRegion1 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 4*(by_gap + bh) + by_shift, bw, bh];
pltRegion2 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 3*(by_gap + bh) + by_shift, bw, bh];
pltRegion3 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 2*(by_gap + bh) + by_shift, bw, bh];
pltRegion4 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 1*(by_gap + bh) + by_shift, 0.4*bw, bh];
pltRegion5 = [bx + 0.5 * (bw + bx_gap) + bx_shift,by + 1*(by_gap + bh) + by_shift, 0.4*bw, bh];
pltRegion6 = [bx + 0 * (bw + bx_gap) + bx_shift,by + 0*(by_gap + bh) + by_shift, 0.4*bw, bh];
pltRegion7 = [bx + 0.5 * (bw + bx_gap) + bx_shift,by + 0*(by_gap + bh) + by_shift, 0.4*bw, bh];
pltRegion8 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 4*(by_gap + bh) + by_shift, bw, bh];
pltRegion9 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 3*(by_gap + bh) + by_shift, bw, bh];
pltRegion10 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 2*(by_gap + bh) + by_shift, bw, bh];
pltRegion11 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 1*(by_gap + bh) + by_shift, 0.4*bw, bh];
pltRegion12 = [bx + 1.5 * (bw + bx_gap) + bx_shift,by + 1*(by_gap + bh) + by_shift, 0.4*bw, bh];
pltRegion13 = [bx + 1 * (bw + bx_gap) + bx_shift,by + 0*(by_gap + bh) + by_shift, 0.4*bw, bh];
pltRegion14 = [bx + 1.5 * (bw + bx_gap) + bx_shift,by + 0*(by_gap + bh) + by_shift, 0.4*bw, bh];
fig = plt.figure();
ax1 = fig.add_axes(pltRegion1);
ax2 = fig.add_axes(pltRegion2);
ax3 = fig.add_axes(pltRegion3);
ax4 = fig.add_axes(pltRegion4);
ax5 = fig.add_axes(pltRegion5);
ax6 = fig.add_axes(pltRegion6);
ax7 = fig.add_axes(pltRegion7);
ax8 = fig.add_axes(pltRegion8);
ax9 = fig.add_axes(pltRegion9);
ax10 = fig.add_axes(pltRegion10);
ax11 = fig.add_axes(pltRegion11);
ax12 = fig.add_axes(pltRegion12);
ax13 = fig.add_axes(pltRegion13);
ax14 = fig.add_axes(pltRegion14);
cList = ['red','blue','green','black','m'];
lList = ['r-','b-','g-','k-','m-'];
ax = ax1;
ax.plot(t, x,'r-');
ax.scatter(t[peaks_x],x[peaks_x],c='blue');
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax.set_title("Original");
ax = ax2;
ax.scatter(t_peak_x, x_peak_interval,s=1,c='red');
ax.set_ylim([0,10]);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('Interval in sec', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax3;
ax.scatter(t_peak_x, x_peak_interval,s=1,c='red');
ax.set_ylim([0,1]);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('Interval in sec', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax4;
ax.scatter(x_peak_interval[1:], np.roll(x_peak_interval,1)[1:],s=1,c='red');
ax.set_xlabel('interval(n)', fontsize = FontSize_Xlabel );
ax.set_ylabel('interval(n+1)', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax5;
ax.scatter(x_peak_interval[1:], np.roll(x_peak_interval,1)[1:],s=1,c='red');
ax.set_xlim([0,1]);
ax.set_ylim([0,1]);
ax.set_xlabel('interval(n)', fontsize = FontSize_Xlabel );
ax.set_ylabel('interval(n+1)', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax6;
ax.scatter(x_peak_interval[2:], np.roll(x_peak_interval,2)[2:],s=1,c='red');
ax.set_xlabel('interval(n)', fontsize = FontSize_Xlabel );
ax.set_ylabel('interval(n+2)', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax7;
ax.scatter(x_peak_interval[2:], np.roll(x_peak_interval,2)[2:],s=1,c='red');
ax.set_xlim([0,1]);
ax.set_ylim([0,1]);
ax.set_xlabel('interval(n)', fontsize = FontSize_Xlabel );
ax.set_ylabel('interval(n+2)', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax8;
ax.plot(t, xf,'b-');
ax.scatter(t[peaks_xf],xf[peaks_xf],c='red');
ax.set_ylim(-4,4);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('ECG in mV', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax.set_title("Filtered");
ax = ax9;
ax.scatter(t_peak_xf, xf_peak_interval,s=1,c='red');
ax.set_ylim([0,10]);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('Interval in s', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax10;
ax.scatter(t_peak_xf, xf_peak_interval,s=1,c='red');
ax.set_ylim([0,1]);
ax.set_xlabel('time in s', fontsize = FontSize_Xlabel );
ax.set_ylabel('Interval in s', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax11;
ax.scatter(xf_peak_interval[1:], np.roll(xf_peak_interval,1)[1:],s=1,c='blue');
ax.set_xlabel('interval(n)', fontsize = FontSize_Xlabel );
ax.set_ylabel('interval(n+1)', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax12;
ax.scatter(xf_peak_interval[1:], np.roll(xf_peak_interval,1)[1:],s=1,c='blue');
ax.set_xlim([0,1]);
ax.set_ylim([0,1]);
ax.set_xlabel('interval(n)', fontsize = FontSize_Xlabel );
ax.set_ylabel('interval(n+1)', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax13;
ax.scatter(xf_peak_interval[2:], np.roll(xf_peak_interval,2)[2:],s=1,c='blue');
ax.set_xlabel('interval(n)', fontsize = FontSize_Xlabel );
ax.set_ylabel('interval(n+1)', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
ax = ax14;
ax.scatter(xf_peak_interval[2:], np.roll(xf_peak_interval,2)[2:],s=1,c='blue');
ax.set_xlim([0,1]);
ax.set_ylim([0,1]);
ax.set_xlabel('interval(n)', fontsize = FontSize_Xlabel );
ax.set_ylabel('interval(n+1)', fontsize = FontSize_Ylabel );
ax.tick_params(axis='both' ,which='major', labelsize = 6);
plt.gcf().set_size_inches(8.0,9);
fig.show();
|
Reference :
[1]
|
|